Finding Ocean Eddies using Satellite Altimetry: Part 1#
In this practical, we will explore how satellite altimetry data can be used to detect ocean eddies. We will work through a step-by-step process to identify a known eddy using a ready-made eddy database (the Mesoscale Eddy Trajectory Atlas), find and download nearby altimetry mesaurements from Sentinel-3, and apply optimal interpolation using GPSat to reconstruct the eddy’s sea surface height anomaly (SSHA).
Mesoscale Eddy Trajectory Atlas#
AVISO+ provides a database of mesoscale eddies created using observations of multiple satellite altimeters over the last 3 decades. In this notebook, we will work with the META3.2 DT dataset to visualise eddy distributions and identify potentially interesting regions where optimal interpolation of our own altimeter-derived SLA measurements could be applied.
Key Features of META3.2 DT:
Temporal Coverage: 1993–2022
Spatial Resolution: Near-global coverage with detailed tracking of individual eddies.
Eddy Parameters: Position, lifespan, rotation type (cyclonic/anticyclonic), size, and height, rotational velocity, etc.
Applications: Ocean circulation studies, climate research, marine biology, and operational oceanography.
Derivation: The META3.2 DT dataset is based on satellite altimetry observations of sea surface height (SSH) collected from multiple satellite missions over the last three decades. Absolute Dynamic Topography (ADT), which combines SSH with geoid corrections, serves as the primary input for eddy detection. A high-pass Lanczos filter (cutoff: 700 km) is applied to the ADT fields to remove large-scale ocean variability, isolating mesoscale eddies in the 100–300 km range.
Eddy boundaries are then identified using closed SSH contours from these filtered ADT maps, and they are classified as cyclonic or anticyclonic based on their rotation direction. The eddies are tracked over time by comparing their effective contours in consecutive daily altimetry-derived SSH fields, ensuring a minimum 5% overlap between detections.
For a detailed description of the detection and tracking methodology, refer to Pegliasco et al. (2022b) (https://doi.org/10.5194/essd-14-1087-2022) and the AVISO+ documentation found at https://www.aviso.altimetry.fr/en/data/products/value-added-products/global-mesoscale-eddy-trajectory-product/meta3-2-dt.html.
Here is a great video by AVISO demonstating the product: https://www.youtube.com/watch?v=dF3-G4RsM5c
Data reference: The altimetric Mesoscale Eddy Trajectory Atlas product (META3.2 DT allsat, DOI: https://10.24400/527896/a01-2022.005.220209; (Pegliasco et al., 2022)) was produced by SSALTO/DUACS and distributed by AVISO+ (https://www.aviso.altimetry.fr/) with support from CNES, in collaboration with IMEDEA. This atlas was downloaded 10-03-2025, and covers the period from January 1993 to January 2022
Let’s explore the data#
! pip install xarray numpy matplotlib netcdf4 cartopy pathlib geopandas folium pyarrow ipywidgets ipympl dask scipy
Requirement already satisfied: xarray in /usr/local/lib/python3.11/dist-packages (2025.1.2)
Requirement already satisfied: numpy in /usr/local/lib/python3.11/dist-packages (1.26.4)
Requirement already satisfied: matplotlib in /usr/local/lib/python3.11/dist-packages (3.10.0)
Collecting netcdf4
Downloading netCDF4-1.7.2-cp311-cp311-manylinux_2_17_x86_64.manylinux2014_x86_64.whl.metadata (1.8 kB)
Collecting cartopy
Downloading Cartopy-0.24.1-cp311-cp311-manylinux_2_17_x86_64.manylinux2014_x86_64.whl.metadata (7.9 kB)
Requirement already satisfied: pathlib in /usr/local/lib/python3.11/dist-packages (1.0.1)
Requirement already satisfied: geopandas in /usr/local/lib/python3.11/dist-packages (1.0.1)
Requirement already satisfied: folium in /usr/local/lib/python3.11/dist-packages (0.19.5)
Requirement already satisfied: pyarrow in /usr/local/lib/python3.11/dist-packages (18.1.0)
Requirement already satisfied: ipywidgets in /usr/local/lib/python3.11/dist-packages (7.7.1)
Collecting ipympl
Downloading ipympl-0.9.7-py3-none-any.whl.metadata (8.7 kB)
Requirement already satisfied: dask in /usr/local/lib/python3.11/dist-packages (2024.12.1)
Requirement already satisfied: scipy in /usr/local/lib/python3.11/dist-packages (1.13.1)
Requirement already satisfied: packaging>=23.2 in /usr/local/lib/python3.11/dist-packages (from xarray) (24.2)
Requirement already satisfied: pandas>=2.1 in /usr/local/lib/python3.11/dist-packages (from xarray) (2.2.2)
Requirement already satisfied: contourpy>=1.0.1 in /usr/local/lib/python3.11/dist-packages (from matplotlib) (1.3.1)
Requirement already satisfied: cycler>=0.10 in /usr/local/lib/python3.11/dist-packages (from matplotlib) (0.12.1)
Requirement already satisfied: fonttools>=4.22.0 in /usr/local/lib/python3.11/dist-packages (from matplotlib) (4.56.0)
Requirement already satisfied: kiwisolver>=1.3.1 in /usr/local/lib/python3.11/dist-packages (from matplotlib) (1.4.8)
Requirement already satisfied: pillow>=8 in /usr/local/lib/python3.11/dist-packages (from matplotlib) (11.1.0)
Requirement already satisfied: pyparsing>=2.3.1 in /usr/local/lib/python3.11/dist-packages (from matplotlib) (3.2.1)
Requirement already satisfied: python-dateutil>=2.7 in /usr/local/lib/python3.11/dist-packages (from matplotlib) (2.8.2)
Collecting cftime (from netcdf4)
Downloading cftime-1.6.4.post1-cp311-cp311-manylinux_2_17_x86_64.manylinux2014_x86_64.whl.metadata (8.7 kB)
Requirement already satisfied: certifi in /usr/local/lib/python3.11/dist-packages (from netcdf4) (2025.1.31)
Requirement already satisfied: shapely>=1.8 in /usr/local/lib/python3.11/dist-packages (from cartopy) (2.0.7)
Requirement already satisfied: pyshp>=2.3 in /usr/local/lib/python3.11/dist-packages (from cartopy) (2.3.1)
Requirement already satisfied: pyproj>=3.3.1 in /usr/local/lib/python3.11/dist-packages (from cartopy) (3.7.1)
Requirement already satisfied: pyogrio>=0.7.2 in /usr/local/lib/python3.11/dist-packages (from geopandas) (0.10.0)
Requirement already satisfied: branca>=0.6.0 in /usr/local/lib/python3.11/dist-packages (from folium) (0.8.1)
Requirement already satisfied: jinja2>=2.9 in /usr/local/lib/python3.11/dist-packages (from folium) (3.1.5)
Requirement already satisfied: requests in /usr/local/lib/python3.11/dist-packages (from folium) (2.32.3)
Requirement already satisfied: xyzservices in /usr/local/lib/python3.11/dist-packages (from folium) (2025.1.0)
Requirement already satisfied: ipykernel>=4.5.1 in /usr/local/lib/python3.11/dist-packages (from ipywidgets) (6.17.1)
Requirement already satisfied: ipython-genutils~=0.2.0 in /usr/local/lib/python3.11/dist-packages (from ipywidgets) (0.2.0)
Requirement already satisfied: traitlets>=4.3.1 in /usr/local/lib/python3.11/dist-packages (from ipywidgets) (5.7.1)
Requirement already satisfied: widgetsnbextension~=3.6.0 in /usr/local/lib/python3.11/dist-packages (from ipywidgets) (3.6.10)
Requirement already satisfied: ipython>=4.0.0 in /usr/local/lib/python3.11/dist-packages (from ipywidgets) (7.34.0)
Requirement already satisfied: jupyterlab-widgets>=1.0.0 in /usr/local/lib/python3.11/dist-packages (from ipywidgets) (3.0.13)
Requirement already satisfied: click>=8.1 in /usr/local/lib/python3.11/dist-packages (from dask) (8.1.8)
Requirement already satisfied: cloudpickle>=3.0.0 in /usr/local/lib/python3.11/dist-packages (from dask) (3.1.1)
Requirement already satisfied: fsspec>=2021.09.0 in /usr/local/lib/python3.11/dist-packages (from dask) (2024.10.0)
Requirement already satisfied: partd>=1.4.0 in /usr/local/lib/python3.11/dist-packages (from dask) (1.4.2)
Requirement already satisfied: pyyaml>=5.3.1 in /usr/local/lib/python3.11/dist-packages (from dask) (6.0.2)
Requirement already satisfied: toolz>=0.10.0 in /usr/local/lib/python3.11/dist-packages (from dask) (0.12.1)
Requirement already satisfied: importlib_metadata>=4.13.0 in /usr/local/lib/python3.11/dist-packages (from dask) (8.6.1)
Requirement already satisfied: zipp>=3.20 in /usr/local/lib/python3.11/dist-packages (from importlib_metadata>=4.13.0->dask) (3.21.0)
Requirement already satisfied: debugpy>=1.0 in /usr/local/lib/python3.11/dist-packages (from ipykernel>=4.5.1->ipywidgets) (1.8.0)
Requirement already satisfied: jupyter-client>=6.1.12 in /usr/local/lib/python3.11/dist-packages (from ipykernel>=4.5.1->ipywidgets) (6.1.12)
Requirement already satisfied: matplotlib-inline>=0.1 in /usr/local/lib/python3.11/dist-packages (from ipykernel>=4.5.1->ipywidgets) (0.1.7)
Requirement already satisfied: nest-asyncio in /usr/local/lib/python3.11/dist-packages (from ipykernel>=4.5.1->ipywidgets) (1.6.0)
Requirement already satisfied: psutil in /usr/local/lib/python3.11/dist-packages (from ipykernel>=4.5.1->ipywidgets) (5.9.5)
Requirement already satisfied: pyzmq>=17 in /usr/local/lib/python3.11/dist-packages (from ipykernel>=4.5.1->ipywidgets) (24.0.1)
Requirement already satisfied: tornado>=6.1 in /usr/local/lib/python3.11/dist-packages (from ipykernel>=4.5.1->ipywidgets) (6.4.2)
Requirement already satisfied: setuptools>=18.5 in /usr/local/lib/python3.11/dist-packages (from ipython>=4.0.0->ipywidgets) (75.1.0)
Collecting jedi>=0.16 (from ipython>=4.0.0->ipywidgets)
Downloading jedi-0.19.2-py2.py3-none-any.whl.metadata (22 kB)
Requirement already satisfied: decorator in /usr/local/lib/python3.11/dist-packages (from ipython>=4.0.0->ipywidgets) (4.4.2)
Requirement already satisfied: pickleshare in /usr/local/lib/python3.11/dist-packages (from ipython>=4.0.0->ipywidgets) (0.7.5)
Requirement already satisfied: prompt-toolkit!=3.0.0,!=3.0.1,<3.1.0,>=2.0.0 in /usr/local/lib/python3.11/dist-packages (from ipython>=4.0.0->ipywidgets) (3.0.50)
Requirement already satisfied: pygments in /usr/local/lib/python3.11/dist-packages (from ipython>=4.0.0->ipywidgets) (2.18.0)
Requirement already satisfied: backcall in /usr/local/lib/python3.11/dist-packages (from ipython>=4.0.0->ipywidgets) (0.2.0)
Requirement already satisfied: pexpect>4.3 in /usr/local/lib/python3.11/dist-packages (from ipython>=4.0.0->ipywidgets) (4.9.0)
Requirement already satisfied: MarkupSafe>=2.0 in /usr/local/lib/python3.11/dist-packages (from jinja2>=2.9->folium) (3.0.2)
Requirement already satisfied: pytz>=2020.1 in /usr/local/lib/python3.11/dist-packages (from pandas>=2.1->xarray) (2025.1)
Requirement already satisfied: tzdata>=2022.7 in /usr/local/lib/python3.11/dist-packages (from pandas>=2.1->xarray) (2025.1)
Requirement already satisfied: locket in /usr/local/lib/python3.11/dist-packages (from partd>=1.4.0->dask) (1.0.0)
Requirement already satisfied: six>=1.5 in /usr/local/lib/python3.11/dist-packages (from python-dateutil>=2.7->matplotlib) (1.17.0)
Requirement already satisfied: notebook>=4.4.1 in /usr/local/lib/python3.11/dist-packages (from widgetsnbextension~=3.6.0->ipywidgets) (6.5.5)
Requirement already satisfied: charset-normalizer<4,>=2 in /usr/local/lib/python3.11/dist-packages (from requests->folium) (3.4.1)
Requirement already satisfied: idna<4,>=2.5 in /usr/local/lib/python3.11/dist-packages (from requests->folium) (3.10)
Requirement already satisfied: urllib3<3,>=1.21.1 in /usr/local/lib/python3.11/dist-packages (from requests->folium) (2.3.0)
Requirement already satisfied: parso<0.9.0,>=0.8.4 in /usr/local/lib/python3.11/dist-packages (from jedi>=0.16->ipython>=4.0.0->ipywidgets) (0.8.4)
Requirement already satisfied: jupyter-core>=4.6.0 in /usr/local/lib/python3.11/dist-packages (from jupyter-client>=6.1.12->ipykernel>=4.5.1->ipywidgets) (5.7.2)
Requirement already satisfied: argon2-cffi in /usr/local/lib/python3.11/dist-packages (from notebook>=4.4.1->widgetsnbextension~=3.6.0->ipywidgets) (23.1.0)
Requirement already satisfied: nbformat in /usr/local/lib/python3.11/dist-packages (from notebook>=4.4.1->widgetsnbextension~=3.6.0->ipywidgets) (5.10.4)
Requirement already satisfied: nbconvert>=5 in /usr/local/lib/python3.11/dist-packages (from notebook>=4.4.1->widgetsnbextension~=3.6.0->ipywidgets) (7.16.6)
Requirement already satisfied: Send2Trash>=1.8.0 in /usr/local/lib/python3.11/dist-packages (from notebook>=4.4.1->widgetsnbextension~=3.6.0->ipywidgets) (1.8.3)
Requirement already satisfied: terminado>=0.8.3 in /usr/local/lib/python3.11/dist-packages (from notebook>=4.4.1->widgetsnbextension~=3.6.0->ipywidgets) (0.18.1)
Requirement already satisfied: prometheus-client in /usr/local/lib/python3.11/dist-packages (from notebook>=4.4.1->widgetsnbextension~=3.6.0->ipywidgets) (0.21.1)
Requirement already satisfied: nbclassic>=0.4.7 in /usr/local/lib/python3.11/dist-packages (from notebook>=4.4.1->widgetsnbextension~=3.6.0->ipywidgets) (1.2.0)
Requirement already satisfied: ptyprocess>=0.5 in /usr/local/lib/python3.11/dist-packages (from pexpect>4.3->ipython>=4.0.0->ipywidgets) (0.7.0)
Requirement already satisfied: wcwidth in /usr/local/lib/python3.11/dist-packages (from prompt-toolkit!=3.0.0,!=3.0.1,<3.1.0,>=2.0.0->ipython>=4.0.0->ipywidgets) (0.2.13)
Requirement already satisfied: platformdirs>=2.5 in /usr/local/lib/python3.11/dist-packages (from jupyter-core>=4.6.0->jupyter-client>=6.1.12->ipykernel>=4.5.1->ipywidgets) (4.3.6)
Requirement already satisfied: notebook-shim>=0.2.3 in /usr/local/lib/python3.11/dist-packages (from nbclassic>=0.4.7->notebook>=4.4.1->widgetsnbextension~=3.6.0->ipywidgets) (0.2.4)
Requirement already satisfied: beautifulsoup4 in /usr/local/lib/python3.11/dist-packages (from nbconvert>=5->notebook>=4.4.1->widgetsnbextension~=3.6.0->ipywidgets) (4.13.3)
Requirement already satisfied: bleach!=5.0.0 in /usr/local/lib/python3.11/dist-packages (from bleach[css]!=5.0.0->nbconvert>=5->notebook>=4.4.1->widgetsnbextension~=3.6.0->ipywidgets) (6.2.0)
Requirement already satisfied: defusedxml in /usr/local/lib/python3.11/dist-packages (from nbconvert>=5->notebook>=4.4.1->widgetsnbextension~=3.6.0->ipywidgets) (0.7.1)
Requirement already satisfied: jupyterlab-pygments in /usr/local/lib/python3.11/dist-packages (from nbconvert>=5->notebook>=4.4.1->widgetsnbextension~=3.6.0->ipywidgets) (0.3.0)
Requirement already satisfied: mistune<4,>=2.0.3 in /usr/local/lib/python3.11/dist-packages (from nbconvert>=5->notebook>=4.4.1->widgetsnbextension~=3.6.0->ipywidgets) (3.1.2)
Requirement already satisfied: nbclient>=0.5.0 in /usr/local/lib/python3.11/dist-packages (from nbconvert>=5->notebook>=4.4.1->widgetsnbextension~=3.6.0->ipywidgets) (0.10.2)
Requirement already satisfied: pandocfilters>=1.4.1 in /usr/local/lib/python3.11/dist-packages (from nbconvert>=5->notebook>=4.4.1->widgetsnbextension~=3.6.0->ipywidgets) (1.5.1)
Requirement already satisfied: fastjsonschema>=2.15 in /usr/local/lib/python3.11/dist-packages (from nbformat->notebook>=4.4.1->widgetsnbextension~=3.6.0->ipywidgets) (2.21.1)
Requirement already satisfied: jsonschema>=2.6 in /usr/local/lib/python3.11/dist-packages (from nbformat->notebook>=4.4.1->widgetsnbextension~=3.6.0->ipywidgets) (4.23.0)
Requirement already satisfied: argon2-cffi-bindings in /usr/local/lib/python3.11/dist-packages (from argon2-cffi->notebook>=4.4.1->widgetsnbextension~=3.6.0->ipywidgets) (21.2.0)
Requirement already satisfied: webencodings in /usr/local/lib/python3.11/dist-packages (from bleach!=5.0.0->bleach[css]!=5.0.0->nbconvert>=5->notebook>=4.4.1->widgetsnbextension~=3.6.0->ipywidgets) (0.5.1)
Requirement already satisfied: tinycss2<1.5,>=1.1.0 in /usr/local/lib/python3.11/dist-packages (from bleach[css]!=5.0.0->nbconvert>=5->notebook>=4.4.1->widgetsnbextension~=3.6.0->ipywidgets) (1.4.0)
Requirement already satisfied: attrs>=22.2.0 in /usr/local/lib/python3.11/dist-packages (from jsonschema>=2.6->nbformat->notebook>=4.4.1->widgetsnbextension~=3.6.0->ipywidgets) (25.1.0)
Requirement already satisfied: jsonschema-specifications>=2023.03.6 in /usr/local/lib/python3.11/dist-packages (from jsonschema>=2.6->nbformat->notebook>=4.4.1->widgetsnbextension~=3.6.0->ipywidgets) (2024.10.1)
Requirement already satisfied: referencing>=0.28.4 in /usr/local/lib/python3.11/dist-packages (from jsonschema>=2.6->nbformat->notebook>=4.4.1->widgetsnbextension~=3.6.0->ipywidgets) (0.36.2)
Requirement already satisfied: rpds-py>=0.7.1 in /usr/local/lib/python3.11/dist-packages (from jsonschema>=2.6->nbformat->notebook>=4.4.1->widgetsnbextension~=3.6.0->ipywidgets) (0.23.1)
Requirement already satisfied: jupyter-server<3,>=1.8 in /usr/local/lib/python3.11/dist-packages (from notebook-shim>=0.2.3->nbclassic>=0.4.7->notebook>=4.4.1->widgetsnbextension~=3.6.0->ipywidgets) (1.24.0)
Requirement already satisfied: cffi>=1.0.1 in /usr/local/lib/python3.11/dist-packages (from argon2-cffi-bindings->argon2-cffi->notebook>=4.4.1->widgetsnbextension~=3.6.0->ipywidgets) (1.17.1)
Requirement already satisfied: soupsieve>1.2 in /usr/local/lib/python3.11/dist-packages (from beautifulsoup4->nbconvert>=5->notebook>=4.4.1->widgetsnbextension~=3.6.0->ipywidgets) (2.6)
Requirement already satisfied: typing-extensions>=4.0.0 in /usr/local/lib/python3.11/dist-packages (from beautifulsoup4->nbconvert>=5->notebook>=4.4.1->widgetsnbextension~=3.6.0->ipywidgets) (4.12.2)
Requirement already satisfied: pycparser in /usr/local/lib/python3.11/dist-packages (from cffi>=1.0.1->argon2-cffi-bindings->argon2-cffi->notebook>=4.4.1->widgetsnbextension~=3.6.0->ipywidgets) (2.22)
Requirement already satisfied: anyio<4,>=3.1.0 in /usr/local/lib/python3.11/dist-packages (from jupyter-server<3,>=1.8->notebook-shim>=0.2.3->nbclassic>=0.4.7->notebook>=4.4.1->widgetsnbextension~=3.6.0->ipywidgets) (3.7.1)
Requirement already satisfied: websocket-client in /usr/local/lib/python3.11/dist-packages (from jupyter-server<3,>=1.8->notebook-shim>=0.2.3->nbclassic>=0.4.7->notebook>=4.4.1->widgetsnbextension~=3.6.0->ipywidgets) (1.8.0)
Requirement already satisfied: sniffio>=1.1 in /usr/local/lib/python3.11/dist-packages (from anyio<4,>=3.1.0->jupyter-server<3,>=1.8->notebook-shim>=0.2.3->nbclassic>=0.4.7->notebook>=4.4.1->widgetsnbextension~=3.6.0->ipywidgets) (1.3.1)
Downloading netCDF4-1.7.2-cp311-cp311-manylinux_2_17_x86_64.manylinux2014_x86_64.whl (9.3 MB)
━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━ 9.3/9.3 MB 11.9 MB/s eta 0:00:00
?25hDownloading Cartopy-0.24.1-cp311-cp311-manylinux_2_17_x86_64.manylinux2014_x86_64.whl (11.7 MB)
━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━ 11.7/11.7 MB 28.4 MB/s eta 0:00:00
?25hDownloading ipympl-0.9.7-py3-none-any.whl (515 kB)
━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━ 515.7/515.7 kB 14.4 MB/s eta 0:00:00
?25hDownloading cftime-1.6.4.post1-cp311-cp311-manylinux_2_17_x86_64.manylinux2014_x86_64.whl (1.4 MB)
━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━ 1.4/1.4 MB 21.4 MB/s eta 0:00:00
?25hDownloading jedi-0.19.2-py2.py3-none-any.whl (1.6 MB)
━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━ 1.6/1.6 MB 26.3 MB/s eta 0:00:00
?25hInstalling collected packages: jedi, cftime, netcdf4, cartopy, ipympl
Successfully installed cartopy-0.24.1 cftime-1.6.4.post1 ipympl-0.9.7 jedi-0.19.2 netcdf4-1.7.2
import xarray as xr
import numpy as np
import pandas as pd
from pathlib import Path
import geopandas as gpd
import matplotlib.pyplot as plt
import cartopy.crs as ccrs
import cartopy.feature as cfeature
from datetime import datetime
import folium
from shapely.geometry import Polygon
import matplotlib.colors as mcolors
import matplotlib.cm as cm
import matplotlib.path as mpath
from shapely.ops import transform
import pyproj
from shapely.geometry import shape
from shapely import wkt
import subprocess
import requests
import json
import shutil
import os
import zipfile
import sys
import json
import ipywidgets as widgets
from shapely.geometry import mapping
from IPython.display import display
import matplotlib.lines as mlines
from google.colab import drive
drive.mount('/content/drive')
Mounted at /content/drive
week8_path = Path('/content/drive/MyDrive/AI4EO/Week 8')
The following code was used to subset the orginal eddy data from AVISO and store+save the information relevant to us in a geodataframe with eddy polygons defined. You do not not need to (/cannot) run this cell, however the subsets created is available are the Week 8 directory and we’ll load them in in a momement.#
#Load the cyclonic eddies
#Cyclonic eddies
ds_cyclonic = xr.open_dataset(week8_path / 'META3.2_DT_allsat_Cyclonic_long_19930101_20220209.nc')
# Subset the data around 2019
ds_cyclonic = ds_cyclonic.sel(obs=slice(30500000, 34100000))
#Create polygons of the eddy outlines
def create_polygons(ds, contour_lat_var, contour_lon_var):
polygons = []
for i in range(ds.sizes['obs']):
if i % 50000 == 0:
print(f'Creating polygon {i} of {ds.dims["obs"]}')
latitudes = ds[contour_lat_var].isel(obs=i).values
longitudes = ds[contour_lon_var].isel(obs=i).values
polygon = Polygon(zip(longitudes, latitudes))
polygons.append(polygon)
return polygons
cyclonic_polygons = create_polygons(ds_cyclonic, 'effective_contour_latitude', 'effective_contour_longitude')
# Create a GeoDataFrame to store the data
cyclonic_gdf = gpd.GeoDataFrame({
"track_number": ds_cyclonic['track'].values ,
"time": ds_cyclonic['time'].values,
'longitude': ds_cyclonic['longitude'].values,
'latitude': ds_cyclonic['latitude'].values,
"amplitude": ds_cyclonic['amplitude'].values,
"effective_height": ds_cyclonic['effective_contour_height'].values,
"inner_height": ds_cyclonic['inner_contour_height'].values,
"geometry": cyclonic_polygons
}, geometry="geometry")
cyclonic_gdf = cyclonic_gdf.set_crs(epsg=4326)
# Crop the data to 2019
cyclonic_gdf = cyclonic_gdf[cyclonic_gdf['time'].dt.year == 2019]
# Save as a parquest file
cyclonic_gdf.to_parquet(week8_path / 'cyclonic_eddies_2019.parquet')
print('Cyclonic eddies subset saved')
#Load the cyclonic eddies
#Cyclonic eddies
ds_cyclonic = xr.open_dataset(week8_path / 'META3.2_DT_allsat_Cyclonic_long_19930101_20220209.nc')
# Subset the data around 2019
ds_cyclonic = ds_cyclonic.sel(obs=slice(30500000, 34100000))
#Create polygons of the eddy outlines
cyclonic_polygons = create_polygons(ds_cyclonic, 'effective_contour_latitude', 'effective_contour_longitude')
# Create a GeoDataFrame to store the data
cyclonic_gdf = gpd.GeoDataFrame({
"track_number": ds_cyclonic['track'].values ,
"time": ds_cyclonic['time'].values,
'longitude': ds_cyclonic['longitude'].values,
'latitude': ds_cyclonic['latitude'].values,
"amplitude": ds_cyclonic['amplitude'].values,
"effective_height": ds_cyclonic['effective_contour_height'].values,
"inner_height": ds_cyclonic['inner_contour_height'].values,
"geometry": cyclonic_polygons
}, geometry="geometry")
cyclonic_gdf = cyclonic_gdf.set_crs(epsg=4326)
# Crop the data to 2019
cyclonic_gdf = cyclonic_gdf[cyclonic_gdf['time'].dt.year == 2019]
# Save as a parquest file
cyclonic_gdf.to_parquet(week8_path / 'cyclonic_eddies_2019.parquet')
print('Cyclonic eddies subset saved')
Load the geodataframes containing our eddy subsets#
anticyclonic_gdf = gpd.read_parquet(week8_path / 'anticyclonic_eddies_2019.parquet')
anticyclonic_gdf.head()
| track_number | time | longitude | latitude | amplitude | effective_height | inner_height | geometry | |
|---|---|---|---|---|---|---|---|---|
| 0 | 708928 | 2019-01-01 | 151.964188 | 51.163441 | 0.0226 | 2.000000e-03 | 0.024 | POLYGON ((151.88 50.84, 152.12 50.81, 152.38 5... |
| 1 | 708928 | 2019-01-02 | 151.962189 | 51.155045 | 0.0223 | 2.000000e-03 | 0.024 | POLYGON ((152.12 50.84, 152.38 50.87, 152.44 5... |
| 2 | 708928 | 2019-01-03 | 151.949387 | 51.167412 | 0.0217 | 2.000000e-03 | 0.022 | POLYGON ((152.12 50.86, 152.27 50.88, 152.38 5... |
| 3 | 708928 | 2019-01-04 | 152.008545 | 51.156200 | 0.0224 | 7.771561e-16 | 0.022 | POLYGON ((152.38 50.85, 152.49 50.88, 152.42 5... |
| 4 | 708928 | 2019-01-05 | 152.002136 | 51.166542 | 0.0211 | 7.771561e-16 | 0.020 | POLYGON ((152.38 50.87, 152.4 50.88, 152.41 51... |
cyclonic_gdf = gpd.read_parquet(week8_path / 'cyclonic_eddies_2019.parquet')
cyclonic_gdf.head()
| track_number | time | longitude | latitude | amplitude | effective_height | inner_height | geometry | |
|---|---|---|---|---|---|---|---|---|
| 0 | 713905 | 2019-01-01 | 99.719543 | -40.455067 | 0.2246 | 7.771561e-16 | -0.224 | POLYGON ((98.82 -41.12, 98.65 -40.88, 98.54 -4... |
| 1 | 713905 | 2019-01-02 | 99.691483 | -40.453773 | 0.2298 | 2.000000e-03 | -0.226 | POLYGON ((98.88 -41.2, 98.74 -41.12, 98.5 -40.... |
| 2 | 713905 | 2019-01-03 | 99.656265 | -40.448448 | 0.2341 | 2.000000e-03 | -0.232 | POLYGON ((98.7 -41.12, 98.62 -40.87, 98.5 -40.... |
| 3 | 713905 | 2019-01-04 | 99.623764 | -40.443928 | 0.2375 | 4.000000e-03 | -0.232 | POLYGON ((98.64 -41.12, 98.58 -40.88, 98.47 -4... |
| 4 | 713905 | 2019-01-05 | 99.594589 | -40.442413 | 0.2379 | 4.000000e-03 | -0.232 | POLYGON ((98.66 -41.12, 98.47 -40.62, 98.49 -4... |
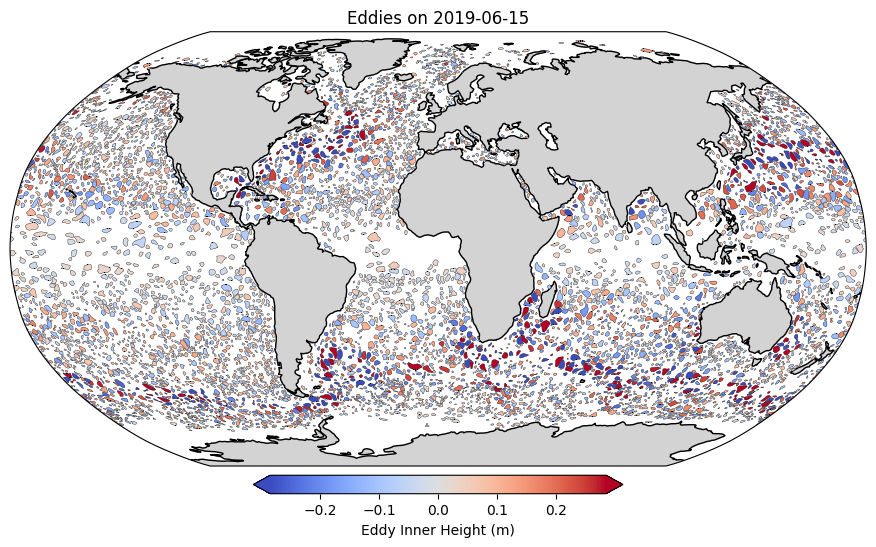
Let’s take a look at the eddies for a single day of interest#
%matplotlib inline
selected_time = "2019-06-15"
selected_time = pd.to_datetime(selected_time)
# Filter eddies for the selected time
date_anticyclonic = anticyclonic_gdf[anticyclonic_gdf["time"] == selected_time]
date_cyclonic = cyclonic_gdf[cyclonic_gdf["time"] == selected_time]
# Plot the eddies on a global map
fig, ax = plt.subplots(figsize=(12, 6), subplot_kw={'projection': ccrs.Robinson()})
# Add features
ax.set_global()
ax.coastlines()
ax.add_feature(cfeature.LAND, color='lightgray')
#get colorscales for inner height
min_mins = min(date_anticyclonic["inner_height"].min(), date_cyclonic["inner_height"].min())
max_maxs = max(date_anticyclonic["inner_height"].max(), date_cyclonic["inner_height"].max())
quartile_mins = min(np.quantile(date_anticyclonic["inner_height"], 0.05), np.quantile(date_cyclonic["inner_height"], 0.05))
quartile_maxs = max(np.quantile(date_anticyclonic["inner_height"], 0.95), np.quantile(date_cyclonic["inner_height"], 0.95))
cmap_bound = max(abs(quartile_mins), abs(quartile_maxs))
norm = mcolors.Normalize(vmin=-cmap_bound, vmax=cmap_bound)
cmap = cm.get_cmap('coolwarm')
for _, row in date_anticyclonic.iterrows():
color = cmap(norm(row["inner_height"]))
ax.add_geometries(
[row.geometry], crs=ccrs.PlateCarree(),
facecolor=color, edgecolor="black", linewidth=0.25,
)
for _, row in date_cyclonic.iterrows():
color = cmap(norm(row["inner_height"]))
ax.add_geometries(
[row.geometry], crs=ccrs.PlateCarree(),
facecolor=color, edgecolor="black", linewidth=0.25,
)
# Add colorbar
sm = cm.ScalarMappable(cmap=cmap, norm=norm)
sm.set_array([])
if min_mins < - cmap_bound and max_maxs > cmap_bound:
extend = "both"
elif min_mins < - cmap_bound:
extend = "min"
elif max_maxs > cmap_bound:
extend = "max"
else:
extend = "neither"
cbar = plt.colorbar(sm, ax=ax, orientation="horizontal", fraction=0.04, pad=0.02, extend=extend)
cbar.set_label("Eddy Inner Height (m)")
# # Plot eddy polygons
# date_anticyclonic.plot(ax=ax, facecolor="red", edgecolor="black", alpha=0.5, transform=ccrs.PlateCarree())
# date_cyclonic.plot(ax=ax, facecolor="blue", edgecolor="black", alpha=0.5, transform=ccrs.PlateCarree())
# Titles and labels
plt.title(f"Eddies on {selected_time.date()}")
plt.show()
<ipython-input-7-ea24c30adc6a>:25: MatplotlibDeprecationWarning: The get_cmap function was deprecated in Matplotlib 3.7 and will be removed in 3.11. Use ``matplotlib.colormaps[name]`` or ``matplotlib.colormaps.get_cmap()`` or ``pyplot.get_cmap()`` instead.
cmap = cm.get_cmap('coolwarm')
/usr/local/lib/python3.11/dist-packages/cartopy/io/__init__.py:241: DownloadWarning: Downloading: https://naturalearth.s3.amazonaws.com/110m_physical/ne_110m_land.zip
warnings.warn(f'Downloading: {url}', DownloadWarning)
/usr/local/lib/python3.11/dist-packages/cartopy/io/__init__.py:241: DownloadWarning: Downloading: https://naturalearth.s3.amazonaws.com/110m_physical/ne_110m_coastline.zip
warnings.warn(f'Downloading: {url}', DownloadWarning)
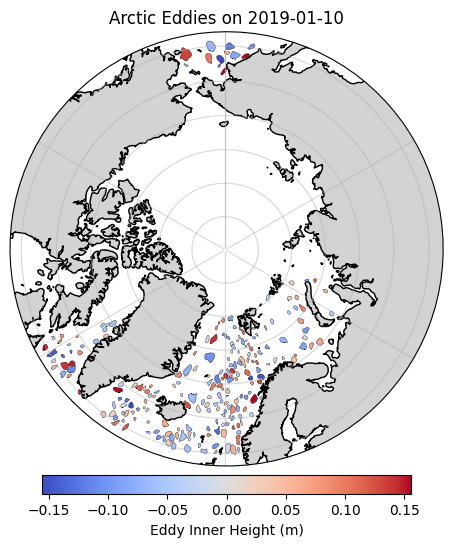
As usual, we’ll be focusing on the Arctic. Let’s crop the dataframe to include only eddies north of 60 N#
arctic_cyclonic_gdf = cyclonic_gdf.cx[0:360, 60:90].copy()
arctic_anticyclonic_gdf = anticyclonic_gdf.cx[0:360, 60:90].copy()
arctic_anticyclonic_gdf.head()
| track_number | time | longitude | latitude | amplitude | effective_height | inner_height | geometry | |
|---|---|---|---|---|---|---|---|---|
| 5504 | 710250 | 2019-01-01 | 345.219666 | 61.521732 | 0.0426 | 0.008 | 0.050 | POLYGON ((345.62 60.81, 345.88 60.79, 346.04 6... |
| 5505 | 710250 | 2019-01-02 | 345.249420 | 61.527199 | 0.0381 | 0.008 | 0.046 | POLYGON ((345.62 60.83, 345.88 60.8, 346.1 60.... |
| 5506 | 710250 | 2019-01-03 | 345.279572 | 61.535881 | 0.0350 | 0.010 | 0.044 | POLYGON ((345.62 60.84, 345.88 60.83, 345.97 6... |
| 5507 | 710250 | 2019-01-04 | 345.287109 | 61.557972 | 0.0330 | 0.014 | 0.046 | POLYGON ((345.88 60.86, 345.91 60.88, 345.8 61... |
| 5508 | 710250 | 2019-01-05 | 345.326294 | 61.552456 | 0.0345 | 0.014 | 0.048 | POLYGON ((345.12 61.09, 345.38 60.97, 345.62 6... |
%matplotlib inline
#Select date
selected_time = "2019-01-10"
# Plot the eddies on a global map for a single day
fig, ax = plt.subplots(figsize=(12, 6), subplot_kw={'projection': ccrs.NorthPolarStereo()})
ax.set_extent([-180, 180, 58, 90], crs=ccrs.PlateCarree())
# Add features
ax.coastlines()
ax.add_feature(cfeature.LAND, color='lightgray')
ax.gridlines(alpha=0.5)
# Get colorscales for inner height
min_mins = min(arctic_anticyclonic_gdf[arctic_anticyclonic_gdf["time"] == selected_time]["inner_height"].min(), arctic_cyclonic_gdf[arctic_cyclonic_gdf["time"] == selected_time]["inner_height"].min())
max_maxs = max(arctic_anticyclonic_gdf[arctic_anticyclonic_gdf["time"] == selected_time]["inner_height"].max(), arctic_cyclonic_gdf[arctic_cyclonic_gdf["time"] == selected_time]["inner_height"].max())
quartile_mins = min(np.quantile(arctic_anticyclonic_gdf[arctic_anticyclonic_gdf["time"] == selected_time]["inner_height"], 0.05), np.quantile(arctic_cyclonic_gdf[arctic_cyclonic_gdf["time"] == selected_time]["inner_height"], 0.05))
quartile_maxs = max(np.quantile(arctic_anticyclonic_gdf[arctic_anticyclonic_gdf["time"] == selected_time]["inner_height"], 0.95), np.quantile(arctic_cyclonic_gdf[arctic_cyclonic_gdf["time"] == selected_time]["inner_height"], 0.95))
cmap_bound = max(abs(quartile_mins), abs(quartile_maxs))
norm = mcolors.Normalize(vmin=-cmap_bound, vmax=cmap_bound)
cmap = cm.get_cmap('coolwarm')
for _, row in arctic_anticyclonic_gdf[arctic_anticyclonic_gdf["time"] == selected_time].iterrows():
color = cmap(norm(row["inner_height"]))
ax.add_geometries(
[row.geometry], crs=ccrs.PlateCarree(),
facecolor=color, edgecolor="black", linewidth=0.25,
)
for _, row in arctic_cyclonic_gdf[arctic_cyclonic_gdf["time"] == selected_time].iterrows():
color = cmap(norm(row["inner_height"]))
ax.add_geometries(
[row.geometry], crs=ccrs.PlateCarree(),
facecolor=color, edgecolor="black", linewidth=0.25,
)
# Add colorbar
sm = cm.ScalarMappable(cmap=cmap, norm=norm)
sm.set_array([])
cbar = plt.colorbar(sm, ax=ax, orientation="horizontal", fraction=0.04, pad=0.02)
cbar.set_label("Eddy Inner Height (m)")
# Make the axis round
theta = np.linspace(0, 2*np.pi, 100)
center, radius = [0.5, 0.5], 0.5
verts = np.vstack([np.sin(theta), np.cos(theta)]).T
circle = mpath.Path(verts * radius + center)
ax.set_boundary(circle, transform=ax.transAxes)
plt.title(f"Arctic Eddies on {selected_time}")
plt.show()
<ipython-input-10-0624be8fefd5>:23: MatplotlibDeprecationWarning: The get_cmap function was deprecated in Matplotlib 3.7 and will be removed in 3.11. Use ``matplotlib.colormaps[name]`` or ``matplotlib.colormaps.get_cmap()`` or ``pyplot.get_cmap()`` instead.
cmap = cm.get_cmap('coolwarm')
/usr/local/lib/python3.11/dist-packages/cartopy/io/__init__.py:241: DownloadWarning: Downloading: https://naturalearth.s3.amazonaws.com/50m_physical/ne_50m_land.zip
warnings.warn(f'Downloading: {url}', DownloadWarning)
/usr/local/lib/python3.11/dist-packages/cartopy/io/__init__.py:241: DownloadWarning: Downloading: https://naturalearth.s3.amazonaws.com/50m_physical/ne_50m_coastline.zip
warnings.warn(f'Downloading: {url}', DownloadWarning)
Eddy Selection#
Option 1: Manual Filtering#
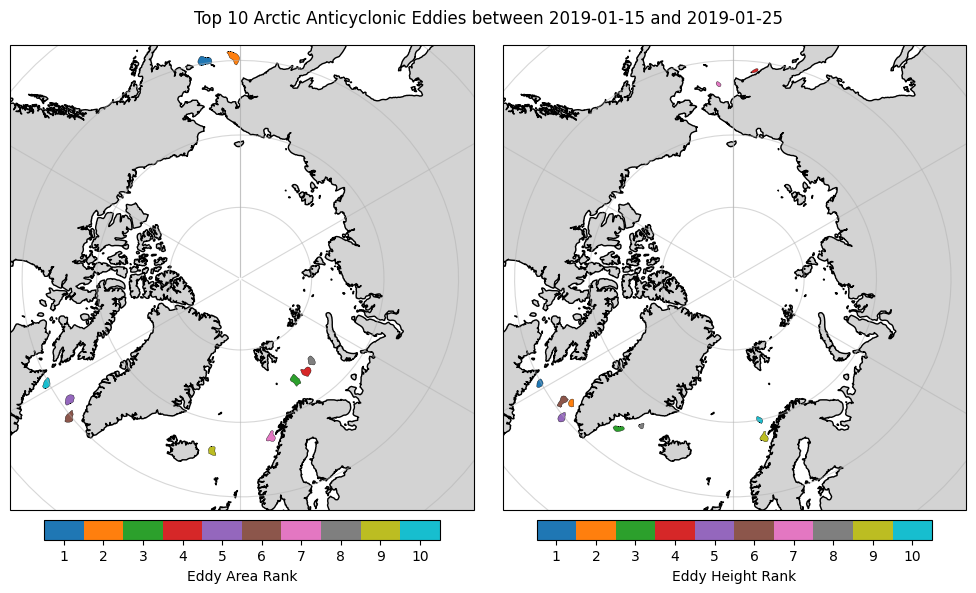
To find interesting eddy to explore later using our own altimery data and optimal interpolation, we could do some simple filtering of the dataframe(s).
#Let's find an interesting anticyclonic eddy based on the time, inner height and/or effective_area
# Find the area of the eddies inside the 'effective contour' (notice we must reproject the geometries to get a more accurate area)
arctic_anticyclonic_gdf["effective_area"] = arctic_anticyclonic_gdf["geometry"].to_crs("EPSG:6931").area
#select the date range
start_date = pd.to_datetime("2019-01-15")
end_date = pd.to_datetime("2019-01-25")
arctic_anticyclonic_gdf_filtered = arctic_anticyclonic_gdf[(arctic_anticyclonic_gdf["time"] >= start_date) & (arctic_anticyclonic_gdf["time"] <= end_date)]
# Let's find the 'big' eddies for possible selection
biggest_areas = arctic_anticyclonic_gdf_filtered.loc[arctic_anticyclonic_gdf_filtered.groupby('track_number')['effective_area'].idxmax()].nlargest(10, "effective_area")
large_heights = arctic_anticyclonic_gdf_filtered.loc[arctic_anticyclonic_gdf_filtered.groupby('track_number')['inner_height'].idxmax()].nlargest(10, "inner_height")
#plot them
%matplotlib inline
# Plot the anticyclonic eddies with the top 10 effective areas and color them by their order
fig, (ax1, ax2) = plt.subplots(1, 2, figsize=(10, 6), subplot_kw={'projection': ccrs.NorthPolarStereo()})
ax1.set_extent([-180, 180, 58, 90], crs=ccrs.PlateCarree())
ax1.coastlines()
ax1.add_feature(cfeature.LAND, color='lightgray')
ax1.gridlines(alpha=0.5)
# Color by order for effective area
norm_area = mcolors.Normalize(vmin=0, vmax=10)
cmap_area = cm.get_cmap('tab10')
for i, (_, row) in enumerate(biggest_areas.iterrows()):
color = cmap_area(norm_area(i))
ax1.add_geometries(
[row.geometry], crs=ccrs.PlateCarree(),
facecolor=color, edgecolor="black", linewidth=0.25,
)
# Add colorbar for effective area
sm_area = cm.ScalarMappable(cmap=cmap_area, norm=norm_area)
sm_area.set_array([])
cbar_area = plt.colorbar(sm_area, ax=ax1, orientation="horizontal", fraction=0.04, pad=0.02)
cbar_area.set_label("Eddy Area Rank")
# Improve colorbar tick labels for effective area
cbar_area.set_ticks(np.linspace(0.5, 9.5, 10))
cbar_area.set_ticklabels([f"{i+1}" for i in range(10)])
# Plot the anticyclonic eddies with the top 10 effective heights and color them by their order
ax2.set_extent([-180, 180, 58, 90], crs=ccrs.PlateCarree())
ax2.coastlines()
ax2.add_feature(cfeature.LAND, color='lightgray')
ax2.gridlines(alpha=0.5)
# Color by order for effective height
norm_height = mcolors.Normalize(vmin=0, vmax=10)
cmap_height = cm.get_cmap('tab10')
for i, (_, row) in enumerate(large_heights.iterrows()):
color = cmap_height(norm_height(i))
ax2.add_geometries(
[row.geometry], crs=ccrs.PlateCarree(),
facecolor=color, edgecolor="black", linewidth=0.25,
)
# Add colorbar for effective height
sm_height = cm.ScalarMappable(cmap=cmap_height, norm=norm_height)
sm_height.set_array([])
cbar_height = plt.colorbar(sm_height, ax=ax2, orientation="horizontal", fraction=0.04, pad=0.02)
cbar_height.set_label("Eddy Height Rank")
# Improve colorbar tick labels for effective height
cbar_height.set_ticks(np.linspace(0.5, 9.5, 10))
cbar_height.set_ticklabels([f"{i+1}" for i in range(10)])
plt.suptitle(f"Top 10 Arctic Anticyclonic Eddies between {start_date.date()} and {end_date.date()}")
plt.tight_layout()
plt.show()
<ipython-input-11-57b0e93f18a6>:28: MatplotlibDeprecationWarning: The get_cmap function was deprecated in Matplotlib 3.7 and will be removed in 3.11. Use ``matplotlib.colormaps[name]`` or ``matplotlib.colormaps.get_cmap()`` or ``pyplot.get_cmap()`` instead.
cmap_area = cm.get_cmap('tab10')
<ipython-input-11-57b0e93f18a6>:55: MatplotlibDeprecationWarning: The get_cmap function was deprecated in Matplotlib 3.7 and will be removed in 3.11. Use ``matplotlib.colormaps[name]`` or ``matplotlib.colormaps.get_cmap()`` or ``pyplot.get_cmap()`` instead.
cmap_height = cm.get_cmap('tab10')
biggest_areas
| track_number | time | longitude | latitude | amplitude | effective_height | inner_height | geometry | effective_area | |
|---|---|---|---|---|---|---|---|---|---|
| 209355 | 721220 | 2019-01-25 | 189.388809 | 59.629944 | 0.0403 | 0.026 | 0.066 | POLYGON ((189.62 58.99, 190.12 58.91, 190.62 5... | 2.044912e+10 |
| 152873 | 719958 | 2019-01-19 | 182.091705 | 59.573280 | 0.0315 | 0.040 | 0.070 | POLYGON ((182.12 58.8, 182.88 58.72, 183.38 58... | 1.901435e+10 |
| 186173 | 720672 | 2019-01-21 | 28.498108 | 73.991829 | 0.0417 | 0.016 | 0.056 | POLYGON ((27.62 73.11, 28.12 73.23, 29.12 73.2... | 1.614833e+10 |
| 177726 | 720481 | 2019-01-22 | 35.991928 | 74.139183 | 0.0536 | 0.014 | 0.066 | POLYGON ((34.38 73.56, 34.88 73.49, 35.73 73.6... | 1.523855e+10 |
| 99623 | 718441 | 2019-01-18 | 305.332520 | 61.162666 | 0.1816 | -0.044 | 0.136 | POLYGON ((304.88 60.59, 305.12 60.45, 305.62 6... | 1.481180e+10 |
| 72681 | 717055 | 2019-01-19 | 309.060699 | 59.647861 | 0.1661 | -0.002 | 0.164 | POLYGON ((308.88 58.97, 309.12 58.97, 309.38 5... | 1.434386e+10 |
| 87759 | 717980 | 2019-01-19 | 10.752258 | 67.765175 | 0.0617 | 0.068 | 0.128 | POLYGON ((10.88 67.08, 11.38 67.04, 11.88 67.1... | 1.366034e+10 |
| 181313 | 720572 | 2019-01-21 | 41.100479 | 74.937454 | 0.0295 | -0.008 | 0.020 | POLYGON ((40.12 74.33, 40.62 74.23, 40.88 74.2... | 1.169499e+10 |
| 101182 | 718498 | 2019-01-19 | 350.651062 | 65.880455 | 0.0233 | -0.010 | 0.012 | POLYGON ((350.62 65.34, 351.12 65.3, 351.62 65... | 1.153702e+10 |
| 170859 | 720315 | 2019-01-19 | 298.268799 | 59.721630 | 0.0716 | 0.132 | 0.202 | POLYGON ((298.38 59.08, 298.62 59.06, 298.88 5... | 1.153048e+10 |
large_heights
| track_number | time | longitude | latitude | amplitude | effective_height | inner_height | geometry | effective_area | |
|---|---|---|---|---|---|---|---|---|---|
| 170855 | 720315 | 2019-01-15 | 298.344696 | 59.793324 | 0.0783 | 1.420000e-01 | 0.220 | POLYGON ((298.62 59.25, 298.88 59.27, 299.09 5... | 8.276140e+09 |
| 192194 | 720827 | 2019-01-24 | 307.399719 | 61.867043 | 0.1640 | 4.600000e-02 | 0.210 | POLYGON ((306.88 61.59, 307.12 61.51, 307.38 6... | 7.423182e+09 |
| 9332 | 711207 | 2019-01-15 | 322.483185 | 63.948158 | 0.1084 | 7.200000e-02 | 0.180 | POLYGON ((321.12 63.57, 321.62 63.48, 322.12 6... | 1.055388e+10 |
| 87571 | 717976 | 2019-01-15 | 174.181198 | 61.193462 | 0.0598 | 1.220000e-01 | 0.180 | POLYGON ((173.38 60.86, 173.62 60.85, 173.88 6... | 3.855151e+09 |
| 72682 | 717055 | 2019-01-20 | 309.020447 | 59.651093 | 0.1564 | 1.400000e-02 | 0.170 | POLYGON ((309.12 59.1, 309.38 59.13, 309.62 59... | 1.124555e+10 |
| 99620 | 718441 | 2019-01-15 | 305.565552 | 61.274590 | 0.1775 | -1.200000e-02 | 0.164 | POLYGON ((305.62 60.19, 305.88 60.21, 306.38 6... | 1.348825e+10 |
| 157869 | 720060 | 2019-01-15 | 184.352264 | 63.020397 | 0.0304 | 1.340000e-01 | 0.164 | POLYGON ((183.88 62.87, 184.12 62.8, 184.38 62... | 3.696855e+09 |
| 53688 | 715854 | 2019-01-16 | 327.964905 | 66.014557 | 0.0591 | 1.040000e-01 | 0.162 | POLYGON ((327.12 65.75, 327.38 65.74, 327.62 6... | 4.893223e+09 |
| 87763 | 717980 | 2019-01-23 | 11.001556 | 67.548401 | 0.0773 | 8.400000e-02 | 0.160 | POLYGON ((10.88 67.04, 11.12 67, 11.62 66.99, ... | 1.187206e+10 |
| 100024 | 718458 | 2019-01-23 | 10.566788 | 70.073784 | 0.1618 | 7.771561e-16 | 0.160 | POLYGON ((10.88 69.52, 11.12 69.56, 11.38 69.6... | 6.529116e+09 |
Make selection
#Based on the maps, I feel like exploring the eddy ranked 5th by effective area
selected_eddy = arctic_anticyclonic_gdf_filtered.loc[biggest_areas.index[5-1]]
selected_eddy
| 99623 | |
|---|---|
| track_number | 718441 |
| time | 2019-01-18 00:00:00 |
| longitude | 305.33252 |
| latitude | 61.162666 |
| amplitude | 0.1816 |
| effective_height | -0.044 |
| inner_height | 0.136 |
| geometry | POLYGON ((304.88 60.59, 305.12 60.45, 305.62 6... |
| effective_area | 14811804544.732437 |
Option 2: Interactive Selection#
We could also choose to browse the eddies in a more interactive style. Move around the figure below and click on some different eddies to see their properties. I will use this viewer to obtain the same eddy as in the previous step.
# Calculate the effective area of the eddies
arctic_cyclonic_gdf["effective_area"] = arctic_cyclonic_gdf["geometry"].to_crs("EPSG:6931").area
arctic_anticyclonic_gdf["effective_area"] = arctic_anticyclonic_gdf["geometry"].to_crs("EPSG:6931").area
#convert the area to km^2
arctic_cyclonic_gdf["effective_area"] = arctic_cyclonic_gdf["effective_area"] / 1e6
arctic_anticyclonic_gdf["effective_area"] = arctic_anticyclonic_gdf["effective_area"] / 1e6
# Ensure CRS is WGS84 (EPSG:4326)
arctic_cyclonic_gdf = arctic_cyclonic_gdf.to_crs(epsg=4326)
arctic_anticyclonic_gdf = arctic_anticyclonic_gdf.to_crs(epsg=4326)
#Add whether cyclonic or anticyclonic
arctic_cyclonic_gdf["type"] = "Cyclonic"
arctic_anticyclonic_gdf["type"] = "Anticyclonic"
# Get available dates
available_dates = sorted(arctic_cyclonic_gdf["time"].dt.date.unique())
# Function to convert GeoDataFrame to GeoJSON
def gdf_to_geojson(gdf):
features = []
for _, row in gdf.iterrows():
feature = {
"type": "Feature",
"geometry": mapping(row.geometry),
"properties": {
"Eddie Type": row["type"],
"Track Number": row["track_number"],
"Time": row["time"].strftime("%Y-%m-%d"),
"Latitude": row.geometry.centroid.y,
"Longitude": row.geometry.centroid.x,
"Area": row["effective_area"],
"Inner Height": row["inner_height"],
},
}
features.append(feature)
return json.dumps({"type": "FeatureCollection", "features": features})
# Function to create Folium map for a selected date
def create_folium_map(selected_date):
selected_date = pd.to_datetime(selected_date)
# Filter eddies for this date
cyclonic_filtered = arctic_cyclonic_gdf[arctic_cyclonic_gdf["time"].dt.date == selected_date.date()]
anticyclonic_filtered = arctic_anticyclonic_gdf[arctic_anticyclonic_gdf["time"].dt.date == selected_date.date()]
print(f"Date: {selected_date}, Cyclonic: {len(cyclonic_filtered)}, Anticyclonic: {len(anticyclonic_filtered)}")
# Create the map
m = folium.Map(location=[0, 180], zoom_start=2, tiles="OpenStreetMap")
# Add Cyclonic Eddies (red)
if not cyclonic_filtered.empty:
folium.GeoJson(
gdf_to_geojson(cyclonic_filtered),
name="Cyclonic Eddies",
popup=folium.GeoJsonPopup(fields=["Eddie Type", "Track Number", "Time", "Latitude", "Longitude", "Area", "Inner Height"],
aliases=["Eddie Type", "Track Number:", "Date:", "Lat:", "Lon:", "Area (m^2):", "Inner Height (m):"]),
tooltip=folium.GeoJsonTooltip(fields=["Eddie Type", "Time", "Latitude", "Longitude", "Area", "Inner Height"],
aliases=["Eddie Type:", "Date:", "Lat:", "Lon:", "Area (km^2):", "Inner Height (m):"]),
color = "blue"
).add_to(m)
# Add Anticyclonic Eddies (blue)
if not anticyclonic_filtered.empty:
folium.GeoJson(
gdf_to_geojson(anticyclonic_filtered),
name="Anticyclonic Eddies",
popup=folium.GeoJsonPopup(fields=["Eddie Type", "Track Number", "Time", "Latitude", "Longitude", "Area", "Inner Height"],
aliases=["Eddie Type", "Track Number:", "Date:", "Lat:", "Lon:", "Area (m^2):", "Inner Height (m):"]),
tooltip=folium.GeoJsonTooltip(fields=["Eddie Type", "Time", "Latitude", "Longitude", "Area", "Inner Height"],
aliases=["Eddie Type:", "Date:", "Lat:", "Lon:", "Area (km^2):", "Inner Height (m):"]),
color = "red"
).add_to(m)
# Add Layer Control
folium.LayerControl().add_to(m)
return m
# Create interactive date slider
date_slider = widgets.SelectionSlider(
options=available_dates,
description="Date",
continuous_update=True,
orientation="horizontal",
layout=widgets.Layout(width="800px")
)
# Output widget to display the map
output = widgets.Output()
# Function to update map when slider is changed
def update_map(change):
with output:
output.clear_output(wait=True) # Clear previous map before rendering a new one
display(create_folium_map(change.new))
# Observe slider changes
date_slider.observe(update_map, names="value")
# Arrange widgets
ui = widgets.VBox([date_slider, output])
# Display UI
display(ui)
# Show initial map
with output:
display(create_folium_map(available_dates[0]))
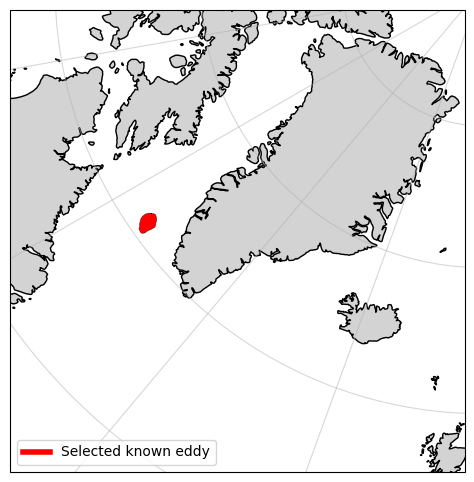
#Having navigated to 2019-01-18, I found the same eddy I was interested in exploring in option 1
#After clicking on it, I see the following information:
eddie_type = "Anticyclonic"
selected_date = "2019-01-18"
selected_track_number = 718441
#I can now use this information to obtain the eddy's data from the dataset
selected_eddy = arctic_anticyclonic_gdf[(arctic_anticyclonic_gdf["time"].dt.date == pd.to_datetime(selected_date).date()) \
& (arctic_anticyclonic_gdf["track_number"] == selected_track_number)].iloc[0]
selected_eddy
| 99623 | |
|---|---|
| track_number | 718441 |
| time | 2019-01-18 00:00:00 |
| longitude | 305.33252 |
| latitude | 61.162666 |
| amplitude | 0.1816 |
| effective_height | -0.044 |
| inner_height | 0.136 |
| geometry | POLYGON ((304.88 60.59, 305.12 60.45, 305.62 6... |
| effective_area | 14811.804545 |
| type | Anticyclonic |
#plot the eddy just to confirm
%matplotlib inline
fig, ax = plt.subplots(figsize=(13, 6), subplot_kw={'projection': ccrs.NorthPolarStereo()})
ax.set_extent([-15, -70, 55, 85], crs=ccrs.PlateCarree())
ax.coastlines()
ax.add_feature(cfeature.LAND, color='lightgray')
ax.gridlines(alpha=0.5)
# Plot the selected eddy
ax.add_geometries(
[selected_eddy['geometry']], crs=ccrs.PlateCarree(),
facecolor="red", edgecolor="black", linewidth=0.25,
)
# Add a legend
eddy_proxy = mlines.Line2D([], [], color='red', linewidth=4, label="Selected known eddy")
ax.legend(handles=[eddy_proxy], loc='lower left')
plt.show()
#Let's save the selected eddy's data to a parquet file
selected_eddy_gdf = gpd.GeoDataFrame([selected_eddy])
selected_eddy_gdf.to_file(week8_path / f"selected_eddy_{selected_eddy['time'].strftime('%Y-%m-%d')}_{selected_eddy['track_number']}.gpkg", layer='row', driver='GPKG')
/usr/local/lib/python3.11/dist-packages/pyogrio/geopandas.py:662: UserWarning: 'crs' was not provided. The output dataset will not have projection information defined and may not be usable in other systems.
write(
Fetching Altimetry Data#
We will now query the Copernicus Dataspace Ecosystem (https://dataspace.copernicus.eu/) and download sentinel-3 altimery data for the selected eddy ± a number of days for optimal interpolation.#
def get_access_token(username, password):
"""
Obtain an access token to the Copernicus Data Space Ecosystem.
Necessary for the download of hosted products.
"""
p = subprocess.run(f"curl --location --request POST 'https://identity.dataspace.copernicus.eu/auth/realms/CDSE/protocol/openid-connect/token' \
--header 'Content-Type: application/x-www-form-urlencoded' \
--data-urlencode 'grant_type=password' \
--data-urlencode 'username={username}' \
--data-urlencode 'password={password}' \
--data-urlencode 'client_id=cdse-public'", shell=True,capture_output=True, text=True)
access_dict = json.loads(p.stdout)
return access_dict['access_token'], access_dict['refresh_token']
#=============================================================================================================================================================#
def get_new_access_token(refresh_token):
"""
Obtain a new access token to the Copernicus Data Space Ecosystem using a previously provided refesh token.
"""
p = subprocess.run(f"curl --location --request POST 'https://identity.dataspace.copernicus.eu/auth/realms/CDSE/protocol/openid-connect/token' \
--header 'Content-Type: application/x-www-form-urlencoded' \
--data-urlencode 'grant_type=refresh_token' \
--data-urlencode 'refresh_token={refresh_token}' \
--data-urlencode 'client_id=cdse-public'", shell=True,capture_output=True, text=True)
access_dict = json.loads(p.stdout)
return access_dict['access_token'], access_dict['refresh_token']
#=============================================================================================================================================================#
def get_S3_SI_search_results_df(date):
"""
Obtain a pandas dataframe of Sentinel-3 sea ice thematic products for a given date.
"""
json = requests.get(f"https://catalogue.dataspace.copernicus.eu/odata/v1/Products?$filter=Collection/Name eq 'SENTINEL-3' \
and Attributes/OData.CSC.StringAttribute/any(att:att/Name eq 'productType' and att/OData.CSC.StringAttribute/Value eq 'SR_2_LAN_SI') \
and Attributes/OData.CSC.StringAttribute/any(att:att/Name eq 'timeliness' and att/OData.CSC.StringAttribute/Value eq 'NT') \
and ContentDate/Start gt {(date-pd.Timedelta(days=1)).strftime('%Y-%m-%dT%H:%M:%SZ')} \
and ContentDate/End lt {(date+pd.Timedelta(days=2)).strftime('%Y-%m-%dT%H:%M:%SZ')}&$top=1000").json()
results_df = pd.DataFrame.from_dict(json['value'])
results_df['Satellite'] = [row['Name'][:3] for i,row in results_df.iterrows()]
results_df['SensingStart'] = [pd.to_datetime(row['ContentDate']['Start']) for i,row in results_df.iterrows()]
results_df['SensingEnd'] = [pd.to_datetime(row['ContentDate']['End']) for i,row in results_df.iterrows()]
results_df = results_df[(results_df['SensingEnd'] >= date) & (results_df['SensingStart'] <= date+pd.Timedelta(days=1))]
results_df['Cycle'] = [row['Name'][-30:-27] for i,row in results_df.iterrows()]
results_df = results_df.sort_values(by='SensingStart')
return results_df
#=============================================================================================================================================================#
def unzip_download(zip_path, save_loc, remove_zip_after_dl=True):
print(f"Unpacking {zip_path.split('/')[-1]}")
shutil.unpack_archive(zip_path, save_loc, 'zip')
# print(f"{zip_path.split('/')[-1]} file unpacked successfully.")
if remove_zip_after_dl == True:
os.remove(zip_path)
#=============================================================================================================================================================#
def is_zipfile_valid(file_path):
try:
with zipfile.ZipFile(file_path, 'r') as zip_ref:
# Check if the ZIP file is valid without extracting its contents
return zip_ref.testzip() is None
except zipfile.BadZipFile:
return False
#=============================================================================================================================================================#
def download_single_product(
product_id, file_name, access_token, download_dir="downloaded_products"
):
"""
Download a single product from the Copernicus Data Space.
:param product_id: The unique identifier for the product.
:param file_name: The name of the file to be downloaded.
:param access_token: The access token for authorization.
:param download_dir: The directory where the product will be saved.
"""
# Ensure the download directory exists
os.makedirs(download_dir, exist_ok=True)
# Construct the download URL
url = (
f"https://zipper.dataspace.copernicus.eu/odata/v1/Products({product_id})/$value"
)
# Set up the session and headers
headers = {"Authorization": f"Bearer {access_token}"}
session = requests.Session()
session.headers.update(headers)
# Perform the request
response = session.get(url, headers=headers, stream=True)
# Check if the request was successful
if response.status_code == 200:
# Define the path for the output file
output_file_path = os.path.join(download_dir, file_name + ".zip")
# Stream the content to a file
with open(output_file_path, "wb") as file:
for chunk in response.iter_content(chunk_size=8192):
if chunk:
file.write(chunk)
print(f"Downloaded: {output_file_path}")
else:
print(
f"Failed to download product {product_id}. Status Code: {response.status_code}"
)
#=============================================================================================================================================================#
def download_S3_from_df(results_df_row, token, target_dir, rm_zip=True):
"""
Download the S3 sea ice thematic corrosponding to a given row in a generated Copernicus Data Space Ecosystem search results dataframe.
S3 sea ice thematic files automatically are stored in the appropriate CPOM server directory.
"""
os.makedirs(target_dir, exist_ok=True)
zip_save_name = results_df_row['Name'][:-5]+'.zip'
zip_path = os.path.join(target_dir,zip_save_name)
# Check we haven't already downloaded + extracted the files
if (os.path.exists(os.path.join(target_dir,results_df_row['Name'],'enhanced_measurement.nc')) == False) or (os.path.exists(os.path.join(target_dir,results_df_row['Name'],'standard_measurement.nc')) == False):
print(f"Downloading {results_df_row['Name'][:-5]}")
# Download the desired product
downloaded_zip_valid = False
while downloaded_zip_valid == False:
# download_product(results_df_row['Id'], zip_path, token, )
download_single_product(results_df_row['Id'], results_df_row['Name'][:-5], token, target_dir)
downloaded_zip_valid = is_zipfile_valid(zip_path)
unzip_download(zip_path, target_dir, rm_zip)
Query Copernicus Dataspace Ecosystem
date = selected_eddy['time']
date_window = pd.Timedelta(days=7)
dates = pd.date_range(date-date_window, date+date_window, freq='D')
#Query the Copernicus Data Space Ecosystem for Sentinel-3 sea ice thematic products for the given date range
search_results = []
for date in dates:
print(f'Querying for {date}')
date = date.tz_localize('UTC')
search_results.append(get_S3_SI_search_results_df(date))
search_results_df = pd.concat(search_results)
print(f'Found {len(search_results_df)} products')
Querying for 2019-01-11 00:00:00
Querying for 2019-01-12 00:00:00
<ipython-input-19-b21795bfd3fb>:48: SettingWithCopyWarning:
A value is trying to be set on a copy of a slice from a DataFrame.
Try using .loc[row_indexer,col_indexer] = value instead
See the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy
results_df['Cycle'] = [row['Name'][-30:-27] for i,row in results_df.iterrows()]
Querying for 2019-01-13 00:00:00
Querying for 2019-01-14 00:00:00
Querying for 2019-01-15 00:00:00
Querying for 2019-01-16 00:00:00
Querying for 2019-01-17 00:00:00
Querying for 2019-01-18 00:00:00
Querying for 2019-01-19 00:00:00
Querying for 2019-01-20 00:00:00
Querying for 2019-01-21 00:00:00
Querying for 2019-01-22 00:00:00
Querying for 2019-01-23 00:00:00
Querying for 2019-01-24 00:00:00
Querying for 2019-01-25 00:00:00
Found 867 products
search_results_df.head()
| @odata.mediaContentType | Id | Name | ContentType | ContentLength | OriginDate | PublicationDate | ModificationDate | Online | EvictionDate | S3Path | Checksum | ContentDate | Footprint | GeoFootprint | Satellite | SensingStart | SensingEnd | Cycle | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 56 | application/octet-stream | 8b0c3868-e35e-37ad-b05c-ba6abb0f89f6 | S3A_SR_2_LAN_SI_20190110T234713_20190111T00075... | application/octet-stream | 44616253 | 2023-10-13T12:44:53.252000Z | 2023-10-19T06:54:14.506975Z | 2023-10-26T16:55:17.961838Z | True | 9999-12-31T23:59:59.999999Z | /eodata/Sentinel-3/SRAL/SR_2_LAN_SI/2019/01/10... | [{'Value': '9d4f3297ddd678407ce214d0de70d2ec',... | {'Start': '2019-01-10T23:47:13.325680Z', 'End'... | geography'SRID=4326;MULTIPOLYGON (((-180 73.19... | {'type': 'MultiPolygon', 'coordinates': [[[[-1... | S3A | 2019-01-10 23:47:13.325680+00:00 | 2019-01-11 00:07:54.433557+00:00 | 040 |
| 57 | application/octet-stream | f320f754-c605-3c0a-917f-9deadb9858c4 | S3B_SR_2_LAN_SI_20190110T235907_20190111T00170... | application/octet-stream | 22961509 | 2023-10-15T16:35:34.756000Z | 2023-10-20T17:01:42.111405Z | 2023-10-20T17:01:43.324661Z | True | 9999-12-31T23:59:59.999999Z | /eodata/Sentinel-3/SRAL/SR_2_LAN_SI/2019/01/10... | [{'Value': '5a08176b18532822cb360a223b97182d',... | {'Start': '2019-01-10T23:59:06.521445Z', 'End'... | geography'SRID=4326;POLYGON ((135.02 -59.9999,... | {'type': 'Polygon', 'coordinates': [[[135.02, ... | S3B | 2019-01-10 23:59:06.521445+00:00 | 2019-01-11 00:17:04.390957+00:00 | 020 |
| 58 | application/octet-stream | 6204cdf9-a284-3dd1-a996-378c2cc19273 | S3A_SR_2_LAN_SI_20190111T003819_20190111T00570... | application/octet-stream | 26258203 | 2023-10-13T12:40:42.891000Z | 2023-10-19T22:08:52.519359Z | 2023-10-19T22:08:54.093727Z | True | 9999-12-31T23:59:59.999999Z | /eodata/Sentinel-3/SRAL/SR_2_LAN_SI/2019/01/11... | [{'Value': '906d8804667a62dcbcbcdb9f6e22a446',... | {'Start': '2019-01-11T00:38:19.237623Z', 'End'... | geography'SRID=4326;POLYGON ((125.867 -59.0213... | {'type': 'Polygon', 'coordinates': [[[125.867,... | S3A | 2019-01-11 00:38:19.237623+00:00 | 2019-01-11 00:57:01.080070+00:00 | 040 |
| 59 | application/octet-stream | 5cb33bb9-d4d3-3721-a9a0-384e6fdc333c | S3B_SR_2_LAN_SI_20190111T004449_20190111T01113... | application/octet-stream | 71281741 | 2023-10-15T16:07:26.083000Z | 2023-10-20T16:28:56.845210Z | 2023-10-20T16:28:58.304268Z | True | 9999-12-31T23:59:59.999999Z | /eodata/Sentinel-3/SRAL/SR_2_LAN_SI/2019/01/11... | [{'Value': '260e877c1d65ada07fc1e8d71f1b3a44',... | {'Start': '2019-01-11T00:44:48.989028Z', 'End'... | geography'SRID=4326;MULTIPOLYGON (((-180 77.92... | {'type': 'MultiPolygon', 'coordinates': [[[[-1... | S3B | 2019-01-11 00:44:48.989028+00:00 | 2019-01-11 01:11:36.535257+00:00 | 020 |
| 60 | application/octet-stream | d29718d6-aa15-34b0-b7c5-74be13ff2dba | S3A_SR_2_LAN_SI_20190111T012416_20190111T01515... | application/octet-stream | 48260686 | 2023-10-13T12:40:23.136000Z | 2023-10-19T06:53:09.215483Z | 2023-10-26T16:54:33.415793Z | True | 9999-12-31T23:59:59.999999Z | /eodata/Sentinel-3/SRAL/SR_2_LAN_SI/2019/01/11... | [{'Value': '07af088a18fd4617ee0f54d033df7152',... | {'Start': '2019-01-11T01:24:16.083297Z', 'End'... | geography'SRID=4326;MULTIPOLYGON (((-180 79.49... | {'type': 'MultiPolygon', 'coordinates': [[[[-1... | S3A | 2019-01-11 01:24:16.083297+00:00 | 2019-01-11 01:51:49.750490+00:00 | 040 |
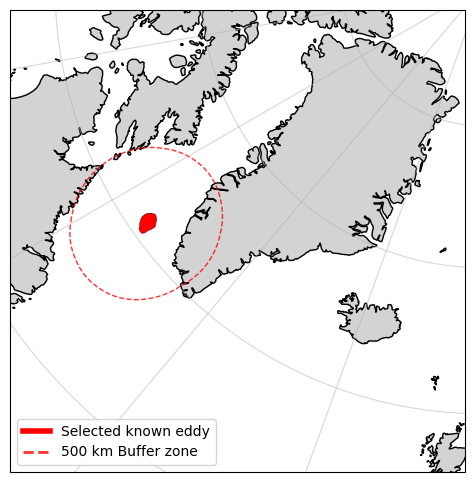
To avoid downloading data that doesn’t pass anywhere near our selected eddy, let us determine which of the query results interects with a defined buffer zone around the eddy
#Now let's find which products intersect with the vicinity of the selected eddy
#Buffer the selected eddy by 500 km
buffer = 500e3
# Convert the selected eddy to a GeoDataFrame and change the CRS to EPSG:6931
# Define the transformation function
project = pyproj.Transformer.from_crs("EPSG:4326", "EPSG:6931", always_xy=True).transform
# Transform the selected eddy geometry to the new CRS
selected_eddy_transformed = transform(project, selected_eddy['geometry'])
# Buffer the transformed geometry
selected_eddy_buffered = selected_eddy_transformed.buffer(buffer)
%matplotlib inline
#plot the selected eddy and the buffered eddy
fig, ax = plt.subplots(figsize=(13, 6), subplot_kw={'projection': ccrs.NorthPolarStereo()})
ax.set_extent([-15, -70, 55, 85], crs=ccrs.PlateCarree())
ax.coastlines()
ax.add_feature(cfeature.LAND, color='lightgray')
ax.gridlines(alpha=0.5)
# Plot the selected eddy
ax.add_geometries(
[selected_eddy['geometry']], crs=ccrs.PlateCarree(),
facecolor="red", edgecolor="black", linewidth=0.25,
)
# Plot the buffered eddy
ax.add_geometries(
[selected_eddy_buffered], crs=ccrs.epsg(6931),
facecolor="none", edgecolor="red", linewidth=1,
linestyle = '--', alpha=0.8
)
# Add a legend
eddy_proxy = mlines.Line2D([], [], color='red', linewidth=4, label="Selected known eddy")
buffer_proxy = mlines.Line2D([], [], color='red', linewidth=2, linestyle='--', alpha=0.8, label=f"{int(buffer/1e3)} km Buffer zone")
ax.legend(handles=[eddy_proxy, buffer_proxy], loc='lower left')
plt.show()
# Convert the GeoFootprint column into shapely geometries
search_results_df["geometry_wkt"] = search_results_df["Footprint"].str.replace(r"geography'SRID=\d+;", "", regex=True)
search_results_df["geometry"] = search_results_df["geometry_wkt"].apply(wkt.loads)
# Convert the DataFrame to a GeoDataFrame
search_results_gdf = gpd.GeoDataFrame(search_results_df, geometry='geometry', crs='EPSG:4326')
#Conver the CRS to EPSG:6931
search_results_gdf = search_results_gdf.to_crs('EPSG:6931')
# Find results that intersect with the buffered eddy
intersecting_products = search_results_gdf[search_results_gdf["geometry"].intersects(selected_eddy_buffered)]
print(f'{len(intersecting_products)} products of {len(search_results_gdf)} in the date range intersect with the selected eddy')
intersecting_products.head()
53 products of 867 in the date range intersect with the selected eddy
| @odata.mediaContentType | Id | Name | ContentType | ContentLength | OriginDate | PublicationDate | ModificationDate | Online | EvictionDate | ... | Checksum | ContentDate | Footprint | GeoFootprint | Satellite | SensingStart | SensingEnd | Cycle | geometry_wkt | geometry | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 59 | application/octet-stream | 5cb33bb9-d4d3-3721-a9a0-384e6fdc333c | S3B_SR_2_LAN_SI_20190111T004449_20190111T01113... | application/octet-stream | 71281741 | 2023-10-15T16:07:26.083000Z | 2023-10-20T16:28:56.845210Z | 2023-10-20T16:28:58.304268Z | True | 9999-12-31T23:59:59.999999Z | ... | [{'Value': '260e877c1d65ada07fc1e8d71f1b3a44',... | {'Start': '2019-01-11T00:44:48.989028Z', 'End'... | geography'SRID=4326;MULTIPOLYGON (((-180 77.92... | {'type': 'MultiPolygon', 'coordinates': [[[[-1... | S3B | 2019-01-11 00:44:48.989028+00:00 | 2019-01-11 01:11:36.535257+00:00 | 020 | MULTIPOLYGON (((-180 77.92870622837371, -177.9... | MULTIPOLYGON (((0 1345599.545, -45679.147 1297... |
| 91 | application/octet-stream | ba576329-e62d-351e-8c43-edf1609ced08 | S3B_SR_2_LAN_SI_20190111T142139_20190111T14383... | application/octet-stream | 38932641 | 2023-10-15T16:44:33.107000Z | 2023-10-20T16:34:00.153352Z | 2023-10-20T16:34:01.467434Z | True | 9999-12-31T23:59:59.999999Z | ... | [{'Value': 'b5b11ecd2a0943d55092e3dcf5fa933a',... | {'Start': '2019-01-11T14:21:39.333210Z', 'End'... | geography'SRID=4326;POLYGON ((84.0104 73.461, ... | {'type': 'Polygon', 'coordinates': [[[84.0104,... | S3B | 2019-01-11 14:21:39.333210+00:00 | 2019-01-11 14:38:35.461506+00:00 | 020 | POLYGON ((84.0104 73.461, 80.192 74.9845, 75.5... | POLYGON ((1830346.809 -192041.304, 1647528.045... |
| 92 | application/octet-stream | 0ea6ab2d-9005-3342-82ce-36b80ba42ff4 | S3A_SR_2_LAN_SI_20190111T150019_20190111T15170... | application/octet-stream | 33443769 | 2023-10-13T12:37:41.200000Z | 2023-10-19T22:02:22.995391Z | 2023-10-19T22:02:24.261230Z | True | 9999-12-31T23:59:59.999999Z | ... | [{'Value': '6303e3c8a8531851be4e84404bf4e9af',... | {'Start': '2019-01-11T15:00:19.014766Z', 'End'... | geography'SRID=4326;POLYGON ((79.1845 70.8485,... | {'type': 'Polygon', 'coordinates': [[[79.1845,... | S3A | 2019-01-11 15:00:19.014766+00:00 | 2019-01-11 15:16:59.936689+00:00 | 040 | POLYGON ((79.1845 70.8485, 76.2546 72.4791, 72... | POLYGON ((2090576.329 -399384.929, 1892952.777... |
| 57 | application/octet-stream | f56b4ac0-4cf4-35a1-96a7-2f46d4157857 | S3B_SR_2_LAN_SI_20190112T001925_20190112T00451... | application/octet-stream | 67206870 | 2023-10-15T16:42:55.297000Z | 2023-10-20T16:27:42.421002Z | 2023-10-20T16:27:43.997860Z | True | 9999-12-31T23:59:59.999999Z | ... | [{'Value': 'b97e1eab37a50a5200a9188e595c8508',... | {'Start': '2019-01-12T00:19:25.239984Z', 'End'... | geography'SRID=4326;MULTIPOLYGON (((-180 76.41... | {'type': 'MultiPolygon', 'coordinates': [[[[-1... | S3B | 2019-01-12 00:19:25.239984+00:00 | 2019-01-12 00:45:10.271717+00:00 | 020 | MULTIPOLYGON (((-180 76.41953418693983, -174.7... | MULTIPOLYGON (((0 1513025.622, -125714.08 1364... |
| 58 | application/octet-stream | 146c0845-5770-312a-860e-de73a74c2d82 | S3A_SR_2_LAN_SI_20190112T005808_20190112T01250... | application/octet-stream | 65541020 | 2023-10-13T12:22:38.096000Z | 2023-10-19T21:59:15.737721Z | 2023-10-19T21:59:18.020499Z | True | 9999-12-31T23:59:59.999999Z | ... | [{'Value': '9720fd77b69f5a81dda89139447e9d40',... | {'Start': '2019-01-12T00:58:08.366518Z', 'End'... | geography'SRID=4326;MULTIPOLYGON (((-180 78.52... | {'type': 'MultiPolygon', 'coordinates': [[[[-1... | S3A | 2019-01-12 00:58:08.366518+00:00 | 2019-01-12 01:25:03.032750+00:00 | 040 | MULTIPOLYGON (((-180 78.52826525575624, -174.8... | MULTIPOLYGON (((0 1279013.505, -105907.052 118... |
5 rows × 21 columns
%matplotlib inline
#plot the search results geometries
fig, ax = plt.subplots(figsize=(13, 6), subplot_kw={'projection': ccrs.NorthPolarStereo()})
ax.set_extent([-15, -70, 55, 85], crs=ccrs.PlateCarree())
ax.coastlines()
ax.add_feature(cfeature.LAND, color='lightgray')
ax.gridlines(alpha=0.5)
# Plot the selected eddy
ax.add_geometries(
[selected_eddy['geometry']], crs=ccrs.PlateCarree(),
facecolor="red", edgecolor="black", linewidth=0.25,
)
# Plot the buffered eddy
ax.add_geometries(
[selected_eddy_buffered], crs=ccrs.epsg(6931),
facecolor="none", edgecolor="red", linewidth=1,
linestyle = '--', alpha=0.8
)
# Plot the search results
intersecting_products.plot(ax=ax, facecolor="none", edgecolor="blue", linewidth=0.5, transform=ccrs.epsg(6931))
# Add a legend
eddy_proxy = mlines.Line2D([], [], color='red', linewidth=4, label="Selected known eddy")
buffer_proxy = mlines.Line2D([], [], color='red', linewidth=2, linestyle='--', alpha=0.8, label=f"{int(buffer/1e3)} km Buffer zone")
s3_proxy = mlines.Line2D([], [], color='blue', linewidth=1, label="Intersecting S3 altimetry tracks")
ax.legend(handles=[eddy_proxy, buffer_proxy, s3_proxy], loc='lower right')
plt.show()
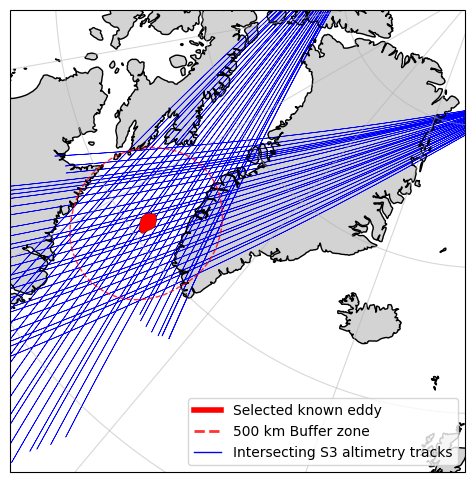
Let’s go ahead and download the relevant S3 altimetry data for our study
target_dir = week8_path / 'S3_SRAL' / f'Eddy_num_{selected_eddy["track_number"]}'
target_dir.mkdir(exist_ok=True, parents=True)
cop_dspace_usrnm = '' # copernicus dataspace username
cop_dspace_psswrd = '' # copernicus dataspace password
token, refresh_token = get_access_token(cop_dspace_usrnm, cop_dspace_psswrd)
for i, row in intersecting_products.iterrows():
download_S3_from_df(row, token, target_dir)
print('Downloaded all products :)')
Downloaded all products :)
We are now ready to use Sentinel-3 A&B altimetry data + optimal interpolation to determine whether we can also retrieve this eddy.
